Exploring microbiome function
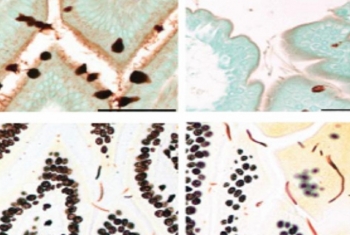
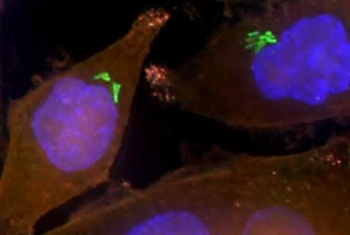
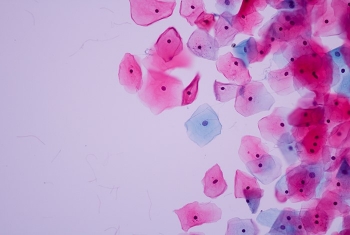
Dr. Shalev Itzkovitz, of the Department of Molecular Cell Biology, has combined RNA sequencing, computational and experimental tools, and advanced single-molecule imaging techniques to uncover interactions among cells, and between cells and the microbiome. Concentrating on the epithelium, the layer of elongated cells that line the intestines, he has discovered that the two sides of the cell differ in the composition of their mRNA and produce different complements of proteins.
Moreover, he found that the number of ribosomes—cellular organelles where proteins are produced—is much higher on the side of the epithelial cells that faces the interior of the gut. The work of Dr. Itzkovitz and his team is helping to clarify how bacteria in the microbiome interact with specialized cell populations and the proteins they produce, thereby enabling the efficient processing of food and the absorption of nutrients.