To support exciting, perhaps risky, innovative ideas in all areas of basic cancer research. The expectation is that this early-phase grant will enable the researcher to obtain enough preliminary results to apply for competitive external funding.
Studying the interplay between EMT and cell invasion in micropatterned stem-cell cultures
Epithelial-to-mesenchymal transition (EMT) and cell migration are pivotal processes in both cancer metastasis and embryonic development and share common molecular and genetic pathways. While EMT has long been considered essential for cancer metastasis, studies have shown that suppressing EMT by deleting key transcription factors, such as Snail or Twist, in primary tumors does not necessarily impair the invasion and metastasis of pancreatic carcinoma cells. A similar phenomenon occurs in embryonic development, where deletion of EMT-related genes leads to gastrulation defects, yet cells can ingress from the epithelial epiblast and migrate across the embryo. This paradox raises a fundamental question: What alternative molecular mechanisms enable cells to exit epithelial tissues in the absence of classical EMT factors? Here, we address this question using a novel micropatterned human pluripotent stem cell model system that captures EMT and cell migration events that occur early in embryonic development. We combine live imaging with CRISPR editing to study the interplay between genetics, EMT, and tissue morphogenesis. This strategy promises to advance our understanding of EMT and cell migration in embryogenesis, with potential implications for providing crucial insights into the mechanisms driving cancer metastasis, paving the way for new therapeutic interventions.

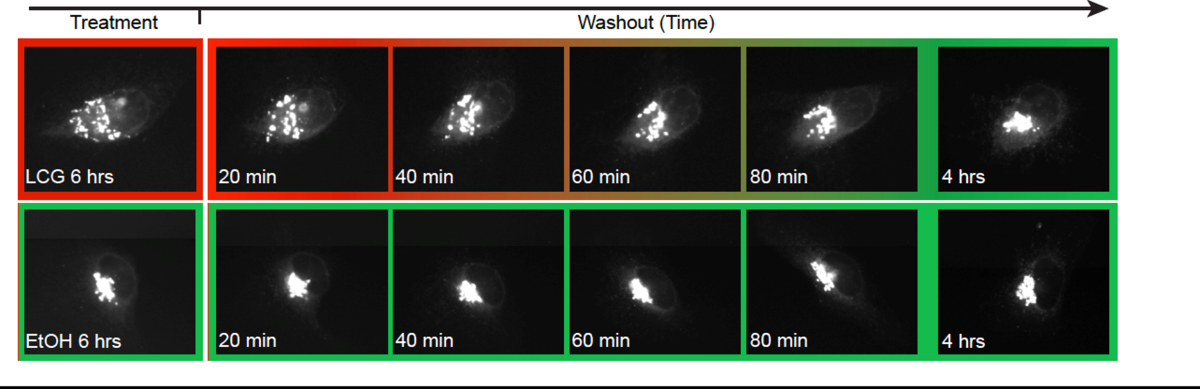
Figure 1. Single-cell tracking using local photoconversion. We apply patterned UV illumination to locally activate a photoconvertible green-to-red fluorescent protein (KikGR). This enables us to track single-cells undergoing EMT and migration in a dense cellular environment
Exploring eIF1AX as a specific vulnerability in loss of chromosome Y (LOY) tumors
Certain types of cancer differentially affect females and males. Recent studies implicated the loss of chromosome Y (LOY) as one of the mechanisms driving male-specific cancers. In these tumors, the loss of paralog Y-linked genes such as eIF1AY creates a dependency on the remaining X-linked eIF1AX paralog gene, which encodes the essential translation initiation factor eIF1A. Likewise, certain female cancers are highly vulnerable to eIF1AX depletion due to the a priori limiting amounts of eIF1A. Thus, targeting eIF1AX presents a potentially precise therapeutic strategy for LOY male tumors or certain female cancers. Additionally, eIF1AX is the only example of a translation initiation factor that is recurrently mutated in cancer. We have previously elucidated the function of eIF1A and its cancer-associated mutants and most recently identified the first eIF1A-specific small molecule inhibitors called 1Ais. In this project, we proposed examining the idea that eIF1AX inhibition by 1Ais is a potentially novel, personalized therapeutic approach against female and male-specific cancers that display rate-limiting amounts of eIF1A. We are currently using CRISPR-Cas9 to mutate the chrY-encoded eIF1AY in the male cancer cell line HCT116 to determine how the loss of eIF1AY influences sensitivity to 1Ais. Likewise, we plan to reduce eIF1AX levels in ovarian cancer cell lines by siRNA or CRISPR-Cas9 to assess the sensitivity to 1Ais. Once these experiments are complete, we will evaluate 1Ais in preclinical xenograft models with LOY. We will also soon start to determine the synergistic effects of 1Ais with DNA-damaging chemotherapies. We anticipate that this study will validate eIF1A as a druggable target in specific types of cancer.
Bacterial Metabolic MRI towards mapping the gut microbiome-cancer axis
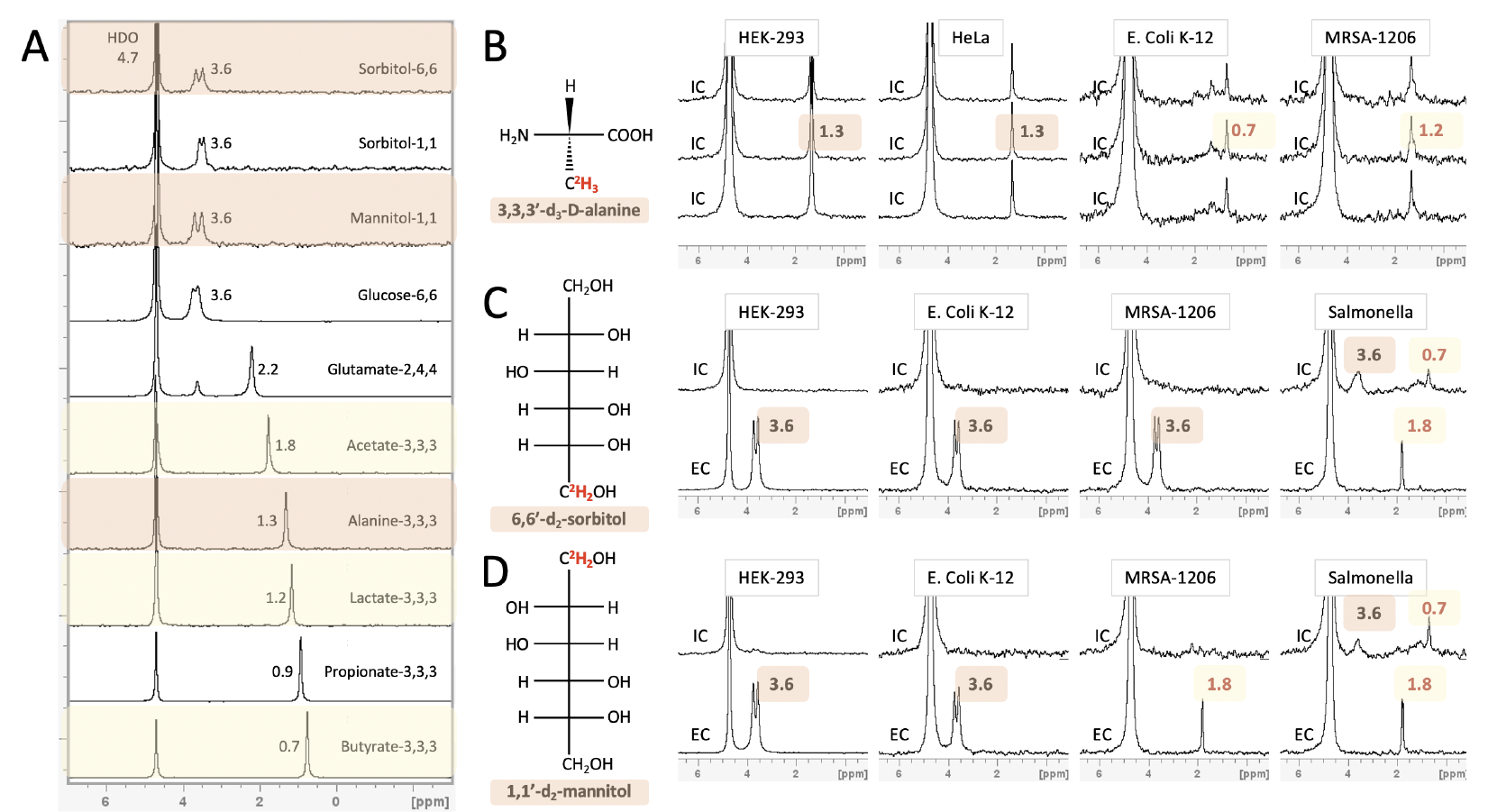
 Despite enormous effort invested in profiling gut microbiota metabolic activity and its relationship to the onset and progression of several cancer types, most microbial metabolic information is obtained from fecal samples that absent bioactive metabolites originating from bacterial populations in the small intestine. In this project, we develop an approach for noninvasive in vivo visualization and mapping gut bacteria's metabolic profiles with MRI. We can obtain spatial maps of downstream metabolites inside an intact subject by rationalizing the labeling of diet components with MRI-detectable isotopes (2H and 19F). We envision a new era of mapping and understanding the gut microbiome-cancer axis, which could be established upon successfully developing this tool.
Despite enormous effort invested in profiling gut microbiota metabolic activity and its relationship to the onset and progression of several cancer types, most microbial metabolic information is obtained from fecal samples that absent bioactive metabolites originating from bacterial populations in the small intestine. In this project, we develop an approach for noninvasive in vivo visualization and mapping gut bacteria's metabolic profiles with MRI. We can obtain spatial maps of downstream metabolites inside an intact subject by rationalizing the labeling of diet components with MRI-detectable isotopes (2H and 19F). We envision a new era of mapping and understanding the gut microbiome-cancer axis, which could be established upon successfully developing this tool.
Role of Regulatory T cells in shaping the tumor microenvironment spatial organization
Regulatory T cells (Tregs) are a critical component of solid tumors, contributing to an immunosuppressive tumor microenvironment (TME). A comprehensive understanding of the mechanisms by which Tregs shape the TME and the factors influencing responses to Treg-targeted therapies remains elusive. Our research focuses on deciphering the complex interactions within the TME and understanding how Tregs influence its spatial organization. In this POC study, we aim to establish methodologies to uncover the intricate web of immune interactions within the TME. Leveraging the PhenoCycler-Fusion System (CODEX), we will perform a spatial characterization of the immune TME of two distinct tumor models, with varying sensitivities to Treg-mediated suppression. Subsequently, we will investigate the impact of Tregs on the spatial organization of the TME. By employing inducible genetic mouse models and Treg-depleting monoclonal antibodies, we will elucidate how Treg elimination influences the immune composition, pathways, and interactions within the TME. This research has the potential to significantly enhance our knowledge of immune surveillance mechanisms and the spatial dynamics of the TME. It may offer new insights into the mechanisms of Treg-mediated tumor immune suppression, potentially optimizing Tregtargeted therapies. Furthermore, we anticipate identifying specific immune cell interactions that could serve as valuable therapeutic targets, paving the way for the development of novel immune cell-based therapies to enhance anti-tumor immunity.
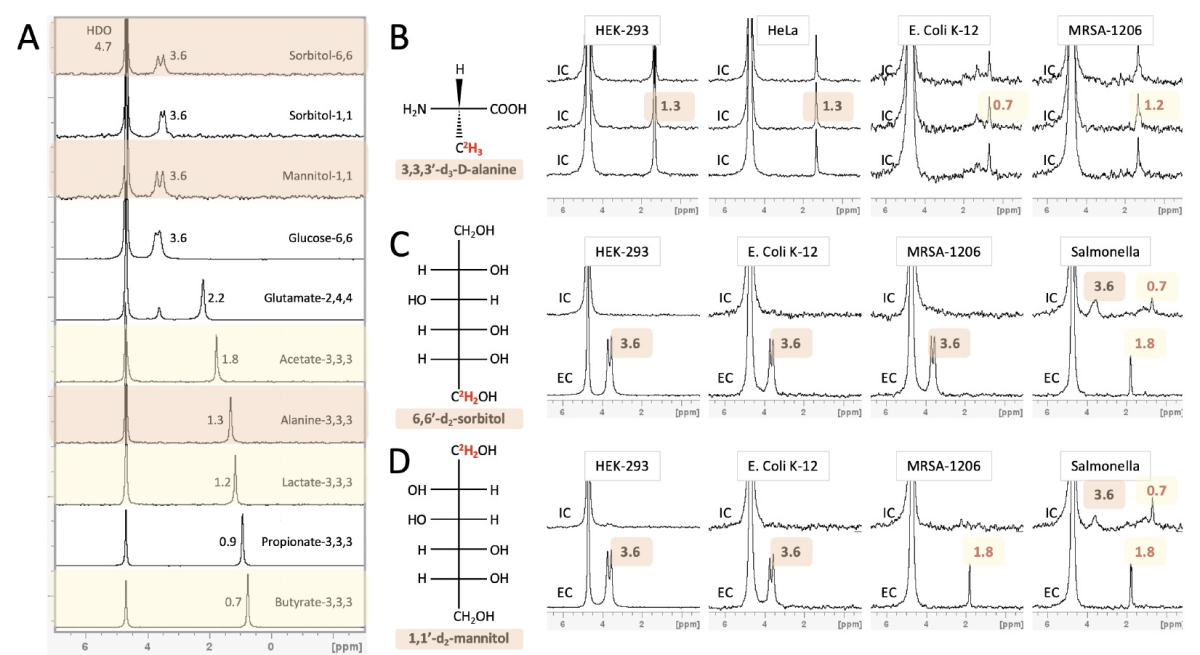
Bacteria-induced translation dysregulation in melanoma as a source for targetable neoantigens
 Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce a durable
Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce a durable
response in patients with metastatic cancer. However, a large fraction of patients do not respond to
immunotherapy. The immune response against tumor cells is driven by neoantigen presentation. Yet, most
tumors harbor a low mutational burden, limiting their targetable neo-peptidomic space. We propose to
address the urgent need to expand the search for neoantigens, by focusing on neopeptides derived from
cancer-specific translation aberrations, an avenue we recently pioneered. We demonstrated that Tryptophan
depletion evokes protein synthesis events that lead to the presentation of immunogenic aberrant peptides.
Moreover, we found an additional class of tumor-presented antigens that are derived from intra-tumor
bacteria that could be used as potential immunotherapy targets.
In this proposal, we suggest utilizing bacteria to deplete tryptophan in the tumor microenvironment by
engineering bacteria that colonize melanoma tumors to overexpress tryptophan-depleting enzymes. We will
assess whether the engineered bacteria induce aberrant peptide presentation by mouse melanoma cells. We
will test in vivo whether the modified bacteria can reach and colonize tumors and cause localized tryptophan
depletion. We will further test how this depletion affects tumor growth and response to checkpoint
inhibitors. Lastly, we will identify presented aberrant peptides in vivo, test the peptides' immunogenicity,
and design mRNA vaccines targeting these peptides. The proposed studies have significant translational
implications, with the potential to identify novel strategies to enhance immunotherapy modalities applicable
to a large number of patients with various tumor mutational burdens.
Deciphering spatial organization of the extracellular matrix in glioblastoma
The extracellular matrix (ECM) is a complex network of proteins and carbohydrates that plays an active role in influencing crucial aspects of cancer biology such as progression, invasion, and drug resistance. We recently established a spatial proteomics approach to map the cell states in glioblastoma at single-cell resolution. We now want to extend this platform to also comprehensively profile the composition and spatial patterns of the ECM. Our goal is to understand the interaction between the ECM, cancer cell states, and non-cancer cells of the tumor microenvironment and how this interplay leads to cancer progression and invasion.
Cancer-associated cachexia
Cancer-associated cachexia is a multifactorial syndrome defined by weight loss and skeletal muscle atrophy, causing decreased quality of life, reduced treatment tolerance, and increased mortality. Although approximately 30% of all cancer deaths are directly caused by cachexia, there is no reliable treatment. We recently demonstrated that an early metabolic rewiring in liver metabolism promoted by the innate immune system contributes to the manifestations of cancer-associated cachexia. Herein, we hypothesize that the liver-brain axis mediated is essential for cachexia development and progression. Thus, we propose that the CNS promotes cachexia by regulating liver metabolism; hence, neuromodulation may effectively deter cachectic progression and improve cancer patients' quality of life.
Exploring microbiome modulators of colorectal cancer signaling pathways
Colorectal cancer (CRC) constitutes one of the leading causes of cancer-related deaths worldwide, which
typically insidiously develops through a long years-long process involving mutations accumulating in
different genes that are essential to a variety of cellular pathways such as WNT, RAS?MAPK, TGF-?, P53,
and DNA mismatch-repair pathways. The gut microbiome has been suggested to causally impact CRC
development, progression, and response to treatment. However, the mechanisms by which gut microbial
signals impact intestinal epithelial cell signaling and transformation remain elusive. Such causal
understanding of host-microbiome modulatory activity on signaling pathways central to CRC development
and clinical behavior may hold promise in harnessing microbes and their secreted products in preventing and
treating CRC. In this proposed project, we aim to establish a high throughput screening pipeline of live
imaging to quantify the simultaneous changes in the major signaling pathways. This will enable us to
massively screen thousands of microbiome-related, food-related and host-related molecules involved in the
host-microbiome niche and their simultaneous effects on multiple key signaling pathways in human CRC.
Top reproducible hits will then be validated on their effects in mouse and human organoid cultures, and then
in in vivo setting in models of CRC in mice (such as AOM/DSS and Apcmin/+). Together, this in vivo to in
vitro to in vivo approach will help in better understanding the host-microbiome modulatory activity on
signaling pathways central to CRC development and clinical behavior, which may hold promise in
harnessing microbes and their secreted products in preventing CRC.
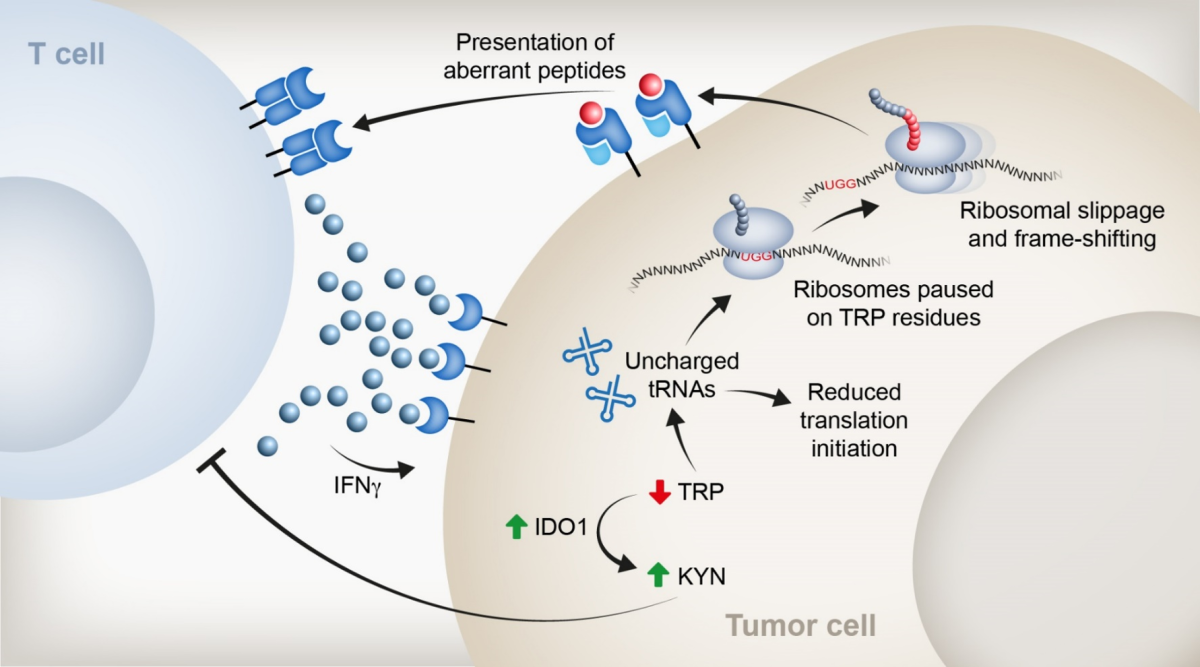
Linking stem cell differentiation processes to cancer initiation
Cancer stem cells (CSCs) are critical drivers of tumor progression and recurrence. Therefore, CSC eradication is crucial for achieving durable remission. However, the molecular pathways leading to CSC induction and accumulation remain poorly understood. We hypothesize that CSCs accumulate due to their inability to give rise to terminally differentiated cells. Our unpublished data demonstrate that under stress conditions, intestinal stem cells (ISCs) differentiate into various lineages of the gut epithelium, potentially as a mechanism to maintain stem cell pool integrity. Our observations suggest that differentiation processes are prioritized to preserve barrier function in tissues required to maintain barrier integrity. We are examining the ability of ISCs to differentiate under stress as a mechanism to ensure the integrity of the stem cell pool. We employ single-cell genomics, transgenic mouse models, and high-resolution imaging techniques to explore the capacity of stem cells to differentiate following DNA damage and eliminate cells with high mutational burden. Furthermore, we aim to demonstrate that the appearance of driver mutations in CRC blocks this differentiation process. This research will advance our understanding of stem cell protective mechanisms that preserve a healthy stem cell pool and prevent cancer initiation. By elucidating these processes, we may identify novel therapeutic targets for cancer prevention and treatment, particularly colorectal cancer.
1. Lineage tracing of intestinal stem cells under irradiation stress. Scheme of the experimental plan (top panel). Image of intestinal crypts in high magnification, showing the labeling of intestinal stem cells with tdTomato upon induction of Tamoxifen for 24h. This system will allow us to follow stem cell differentiation under normal or irradiation stress.
Exploiting autoantibodies for cancer immunotherapy
Malignant tumors present a distinct challenge to the immune system as "altered self". Although there is a well-established crosstalk between the immune system and the tumor that has substantial implications for cancer therapy, the nature of the antigens that allow the immune system to distinguish cancer cells from non-cancer cells has long remained obscure. Recent findings provide an important scientific linkage between the cancer immunology and autoimmunity fields and suggest that targeted breakdown of self-tolerance to various tissue-specific antigens (which are highly expressed by the corresponding tumor) could potentially be exploited as the "magic bullet" to ultimately defeat cancer. It is well established that Aire-deficient mice on NOD genetic background develop severe autoimmune phenotype, characterized by generation of a plethora of autoantibodies against multiple tissues. Therefore, in this research project we aim at exploiting breakdown of immunological tolerance to hundreds of tumor-specific antigens, which occurs naturally in AIRE-deficient mice for cancer immunotherapy. Specifically, we aim at isolating autoantibodies from AIRE-deficient mice, targeting extracellular domains of various membrane-bound antigens associated with various solid tumors, including liver (ASGR1), ovarian (MUC16) and urinary bladder (UPK2) tumors. To this end, we build on our promising preliminary data and experimental platforms we have recently developed for isolation, cloning humanization and expression of autoantibodies against the desired targets. We have identified and isolated several promising autoantibody clones to the above tumor-associated antigens and are in the process of their validation and characterization. The most promising clones will then be used for generation of antibody drug conjugates (ADC), which will then be tested for their tumor cytotoxic capacity in both in vitro and in-vivo mouse xenograft models. So far, we have successfully generated and tested ADC derived from 2B5 mAb clones that specifically targets a unique epitope of MUC16. We hope to generate ADCs for ASGR1 and UPK2 in the near future.
Unraveling the Golgi Stress Response in Cancer and Its Implications for Anti-Tumor Immunity
Dysregulated Golgi homeostasis has emerged as a potential contributor to tumor development and progression by altering the surface and secreted proteome, as well as Golgi-specific modifications. In this proposal we aim to uncover the effects of Golgi stress and perturbed Golgi morphology on cancer cell behavior and their interactions with the immune cells. Our research objectives encompass three specific aims: First, characterizing Golgi stress-induced Golgi fragmentation in a diverse panel of cancer cell lines, shedding light on the mechanisms underlying this phenomenon. Second, investigating the influence of Golgi stress on the surface and secreted proteomes of cancer cells, identifying pivotal effector proteins involved in mediating these alterations. Third, evaluating whether and how Golgi-related aberrations can disrupt interactions between cancer cells and immune cells in in vitro settings, offering insights into the intricate dynamics of tumor-immune crosstalk. Altogether, this comprehensive investigation into the role of Golgi stress in shaping cancer cell phenotype, proteome composition, and immune interactions promises to provide novel insights into tumor-host relationships and may uncover novel therapeutic strategies that harness the Golgi apparatus as a potential target to improve cancer immunotherapy outcomes.
Merging tumor transcriptomic and small extracellular vesicles proteomic to predict breast cancer prognosis, heterogeneity, and therapeutic response
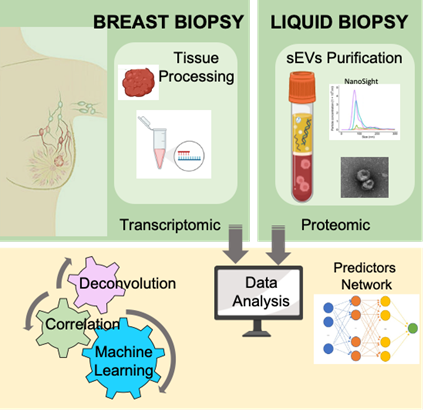 Liquid biopsy is a promising non-invasive approach for early detection of cancer and monitoring disease progression and therapeutic response. We have recently developed a powerful and reliable method to isolate circulating small extracellular vesicles (sEVs) from plasma of breast cancer patients and showed that semi-quantitative proteomic profiling of sEVs can be used for early detection of breast cancer. Our preliminary analysis also suggests that the sEVs proteome could be used for prediction of recurrent disease. In this proof-of-concept project, we proposed not only to improve our isolation and profiling methods of circulating sEVs, but also to identify possible connections between the transcriptome of the primary breast tumors and the proteome of the circulating sEVs. To this end, we plan to profile matching samples of sEVs and primary tumors from breast cancer patients and to establish prediction models by machine learning as well as connectivity map between the tumor transcriptome and the sEVs proteome using computational methods. Deconvolution models will be applied to stratify sEVs based on their cells of origin. This combined analysis may predict sEVs biomarkers for distinct properties of the primary tumors, including tumor stage, tumor composition, heterogeneity, and response to treatment.
Liquid biopsy is a promising non-invasive approach for early detection of cancer and monitoring disease progression and therapeutic response. We have recently developed a powerful and reliable method to isolate circulating small extracellular vesicles (sEVs) from plasma of breast cancer patients and showed that semi-quantitative proteomic profiling of sEVs can be used for early detection of breast cancer. Our preliminary analysis also suggests that the sEVs proteome could be used for prediction of recurrent disease. In this proof-of-concept project, we proposed not only to improve our isolation and profiling methods of circulating sEVs, but also to identify possible connections between the transcriptome of the primary breast tumors and the proteome of the circulating sEVs. To this end, we plan to profile matching samples of sEVs and primary tumors from breast cancer patients and to establish prediction models by machine learning as well as connectivity map between the tumor transcriptome and the sEVs proteome using computational methods. Deconvolution models will be applied to stratify sEVs based on their cells of origin. This combined analysis may predict sEVs biomarkers for distinct properties of the primary tumors, including tumor stage, tumor composition, heterogeneity, and response to treatment.
HOW TO IMPROVE CANCER TREATMENT OUTCOMES BY TEACHING PATIENTS?
 In cancer care, patients undergoing chemotherapy are required to manage their illness independently. Such management involves following the doctor's instructions and making critical decisions that affect their survival, such as reporting side effects or going to the emergency room. This research aimed to identify the types of knowledge patients need to manage their symptoms and side effects effectively. We focused on two types of knowledge: a) procedural knowledge, which involves understanding the steps to follow in various scenarios, and b) mechanistic knowledge, which pertains to understanding the underlying processes behind these procedures.
In cancer care, patients undergoing chemotherapy are required to manage their illness independently. Such management involves following the doctor's instructions and making critical decisions that affect their survival, such as reporting side effects or going to the emergency room. This research aimed to identify the types of knowledge patients need to manage their symptoms and side effects effectively. We focused on two types of knowledge: a) procedural knowledge, which involves understanding the steps to follow in various scenarios, and b) mechanistic knowledge, which pertains to understanding the underlying processes behind these procedures.
Our findings revealed that while procedural knowledge was strongly linked to self-efficacy in managing symptoms, mechanistic knowledge was closely related to the patient’s confidence in managing interactions with their doctor for decision-making. That is, both types of knowledge are crucial for optimal disease management. We compared the changes in these knowledge types between a group receiving only traditional training given in the hospital and a group using a learning module that provided mechanistic information about the treatment and disease. We found no improvement in knowledge for the group with traditional training, whereas the group using the learning module showed significant improvements in both types of knowledge. These results highlight the critical role of mechanistic knowledge, which is often overlooked in patient guidance.
Development of a New Diagnostic Modality for Pancreatic Ductal Adenocarcinoma (PDAC)
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy, often diagnosed in advanced and inoperable stage. Diagnosis requires histologic confirmation, typically obtained through endoscopic ultrasound (EUS) and endoscopic retrograde cholangiopancreatography (ERCP). These procedures are invasive and pose significant risks to patients. Recent discoveries in our lab have shown that shed cells from luminal washes carry information about their tissue of origin. This insight surpasses traditional biopsies in detecting conditions like inflammatory bowel disease (IBD). This project aims to develop a new, less-invasive diagnostic approach for PDAC diagnosis. We plan to use the transcriptomic profiles of shed cells to identify PDAC, thus improving patient care by reducing the invasiveness of the diagnostic process.
Unlocking the immune dynamics of metastatic breast cancer
Cancer metastasis is a process in which cancer cells disseminate from the primary tumor, settle and grow at a site other than the primary tumor site. Cancer mortality primarily stems from metastatic recurrence, emphasizing the urgent need for developing effective metastasis-targeted immunotherapies. Metastatic breast cancer still accounts for a substantial portion of breast cancer-related deaths, with a median survival time of about 12 months after metastasis. Understanding the mechanisms of metastasis can enable the development of targeted therapies that may improve survival rates and reduce the burden of the disease. Prof. Ido Amit, in collaboration with Prof. Neta Erez of Tel Aviv University’s Faculty of Medicine, used his state-of-the-art single-cell technologies and a model of metastatic breast cancer to investigate decipher the cellular and molecular events that allow primary breast tumors to spread to the lungs. The team’s findings, published in the journal Cancer Discovery, demonstrate how the spread of breast cancer to the lungs is associated with specific changes in the balance of immune cell populations. As metastasis commences, the lungs experience heightened infiltration of immune cells known as macrophages. The these macrophages are hijaked by the tumor and activate diverse immune suppressive pathways commentating ind dramatic reduction in the anti-cancer activity of the immune system. The team also used advanced spitial technologies to map out where these cell-cell communications and found that a critical sub-population of these regulatory macrophages which activate a unique immune inhibitory signaling pathway (TREM2) is located at the invasive “boundaries” of cancer and shields the tumor from the body's immune defenses. These discoveries can contribute to improved immunotherapy protocols and the development of novel therapies that can save lives.
Interferon Regulatory Factor 1 (IRF1) to Improve Immune-Checkpoint Blockade in the Treatment of Metastatic Melanoma
In this study we found that localized delivery of a type I Interferon (IFNβ) to a severe mouse melanoma model (B16F10) by injecting an adeno-associated-virus (AAV) as delivery system directly into the tumor we sensitize animals to respond favorably to anti-PDL1 immunotherapy. Localized AAV-IFNβ delivery results in enrichment of immune cells in the tumor microenvironment. This may supplement immune recognition of tumors by IFNg, which we show to act synergistically with IFNb. Interestingly, we show that the transcription factor Interferon-Regulatory-Factor 1 (IRF1) mimics the activation by IFNg, and IFNb. This even in the absence of a functional interferon system. These novel findings open the possibility for a new melanoma treatment regime, which combines the activities of IFNg, IFNb and IRF1 for as a more potent melanoma treatment.
Mechanisms driving gene amplification in cancers with wild type p53
Exploring a New Strategy for Anticancer Therapy: Could an Exceptionally Small, Multifunctional, and Multivalent Targeted Drug Delivery System be created from a Single, Unimolecular Component?
Redox Regulation of Tumor-Associated Sialyltransferases
Detection of autoreactive T follicular helper cells in ovarian carcinoma patients
DNA repair Biomarkers for Predicting Response of Lung Cancer Patients to Immunotherapy
Studying Non-Canonical Expression of IGF2 and its Role in Tumorigenesis
The role of stromal heat shock factor 1 (HSF1) in exosome biogenesis and delivery in the tumor microenvironment
Targeting antigen specific CTLs to pulmonary metastatic lesions
Does the failure in innate immunity establish senescence? The c-Abl story
DNA damage response determines cell fate. A classical view is that under DNA damage cells either die or survive while maintaining a certain degree of genome instability and rearrangements. A significant number of these surviving cells harbor MNi. MNi has lately been reported to activate the cGAS-STING innate immunity pathway. Remarkably, the same cGAS-STING pathway plays a role in inducing senescence Our preliminary data suggest that under c-Abl inhibition/knockout the IFN-I pathway is compromised whereas the senescence program is activated. We will investigate the possibility that failure to activate innate immunity (IFN-I) facilitates senescence.
High content CyTOF-based screening for discovery of novel immune-receptor inhibitors for lymphoma treatment (CyTOF-screen)
B cells and B cell lymphoma depend on BCR signaling for their survival. Removal or inhibition of the immune receptor signaling lead to elimination of the malignant cell. Thus, immune-receptor signaling pathways are an excellent target for cancer treatment. We aim to establish a large-scale screening method that generates numerous readouts in response to hundreds of perturbations by combing high throughput screening with CyTOF readouts. This would allow to define the effect of each treatment on immune cells and lymphoma cells from patients and examine multiple parameters readout by CyTOF.
IgG Fc glycome as a checkpoint for antitumor immunity
Anti-tumor antibodies are being produced in patients with different cancer types. Yet, they fail to mediate effective clearance of the tumor cells and to engage efficient anti-tumor immune response. We hypothesize that the Fc-glycan repertoire is a key determinant that regulate the anti-tumor antibody response. Here, we propose to develop mass-spectrometry based assay to characterize the IgG Fc-glycome in patients and mouse models of different cancer types.
Growing to Die – Diverting Cell Size Regulation Mechanisms to Death Pathways in Cancer Cells
We hypothesize that mechanisms underlying cell size regulation in normal cells are diverted to survival/death pathways in cancer cells, raising the prospect of selectively targeting such mechanisms as a therapeutic strategy. Identifying key hubs in intracellular networks that may shift a size-sensing mechanism to control of proliferation or survival is likely to open new avenues for therapeutic development in the future.
Developing an approach for precision treatment of uveal and thyroid cancers
Somatic mutations in eIF1A, especially in the N-terminal tail (NTT), were recently found to be associated with uveal, thyroid and ovarian cancers. Presently very little is known about the biological function of eIF1A in mammalian cells and how perturbations of its function by the mutations contribute to cancer development. We seek to develop pharmacological tools to selectively interfere with the scanning promoting activity of eIF1A, as a novel personalized therapeutic approach against uveal and thyroid cancers bearing the aforementioned gain-of-function mutations in eIF1A.
Intercellular mRNA transfer and the tumor microenvironment
The tumor microenvironment affects the survival, development, and progression of cancer. Particularly, it is thought that the transfer of full length mRNA molecules between cancer and normal cells might profoundly affect the local microenvironment. We recently discovered a phenomenon by which full-length mRNAs can be transferred between cells. In this project we will explore this process in relation to the tumor microenvironment and cell physiology by determining the biological consequences of the transfer of oncogenic or tumor suppressive mRNAs on the phenotype of downstream acceptor cells and determine whether mRNA transfer occurs in vivo employing human tumor xenografts. Lastly, we will use biochemical/genetic approaches to screen for genes involved in mRNA transfer.
Boosting anti-cancer immunotherapy using pH-switchable reagents
Cancer cells tend to acidify their immediate microenvironment due to increased consumption of glucose at limiting oxygen conditions. These unique biophysical properties could be utilized to achieve differential recognition of antigens using the same therapeutic mAbs. Thus, incorporating pH-switchable recognition elements into therapeutic antibodies could result with the desired differential recognition. This would allow increasing the dose of the mAbs to maximize the therapeutic effect while reducing the severity of, or completely eliminate the off-target side effects.
Real time microscopy of the lung vasculature for dissecting the role of the nuclear lamina in tumor cell extravasation and metastasis
Multiple types of circulating tumor cells (CTCs) metastasize into the lung parenchyma by extravasating the pulmonary vasculature. The space between the alveolus and the capillary (approximately 2 um) imposes a significant mechanical barrier on extravasating CTCs that need to squeeze their bulky and stiff nuclei through it with minimal nuclear damage. Using live imaging of the lung parenchyma with 2-photon microscopy set-up integrated with a thoracic window apparatus we will dissect the mechanisms by which cells transiently reduces their nucleus stiffening enabling extravasation. Genetic manipulations of several breast cancer cell lines by transient and permanent lamin A/C silencing and by CRISPR technology to eliminate a key laminin serine phosphorylation site will be utilized to unravel molecular players involved.
Finding the Achilles heel(s) of triple negative breast cancer
Breast cancer is the most prevalent and lethal cancer in women. 15% of breast cancers do not overexpress the estrogen, progesterone or HER2 receptors also known as Triple Negative Breast Cancers (TNBC). This type of cancers is more aggressive, has poor prognosis, and lack any targeted therapy. This stems partly from our lack of understanding of driver events in TNBC and for lack of validated targets for its pathogenicity. We propose to identify new molecular drivers of TNBC and their respective pathways via covalent fragment phenotypic screening, utilizing an innovative electrophilic fragment library. We have assembled a collection of 993 electrophilic fragments for cellular phenotypic screening that will be used to discover proteins that selectively contribute to TNBC viability, validate them orthogonally via knockdowns and explore their biological roles in TNBC pathogenicity. We will harness the covalent nature of these compounds to pull down their target proteins and characterize them via proteomics. This library can offer an unprecedented window to the working of TNBCs.
Tumor infiltrating B lymphocytes: tumor suppressors or just bystanders?
The importance of T cells in antitumor responses is currently well recognized; however, the role of B cells in cancer immunology is only scantly probed and studied. This is presumably due to the lack of reliable models for functional analysis of distinct B cell subsets in the tumor microenvironment and their contribution to both pro- as well as antitumor immune responses. Focusing on pancreatic cancer, we wish to characterize the potential contribution of B-cells in the microenvironment to cancer control, management and therapy. Specifically, we propose to utilize novel intravital imaging modalities integrated with lineage specific B cell reporter mice to study tumor-infiltrating lymphocytes (TIL) B cells in the tumor microenvironment in both mouse and human pancreatic tumors.

 Despite enormous effort invested in profiling gut microbiota metabolic activity and its relationship to the onset and progression of several cancer types, most microbial metabolic information is obtained from fecal samples that absent bioactive metabolites originating from bacterial populations in the small intestine. In this project, we develop an approach for noninvasive in vivo visualization and mapping gut bacteria's metabolic profiles with MRI. We can obtain spatial maps of downstream metabolites inside an intact subject by rationalizing the labeling of diet components with MRI-detectable isotopes (2H and 19F). We envision a new era of mapping and understanding the gut microbiome-cancer axis, which could be established upon successfully developing this tool.
Despite enormous effort invested in profiling gut microbiota metabolic activity and its relationship to the onset and progression of several cancer types, most microbial metabolic information is obtained from fecal samples that absent bioactive metabolites originating from bacterial populations in the small intestine. In this project, we develop an approach for noninvasive in vivo visualization and mapping gut bacteria's metabolic profiles with MRI. We can obtain spatial maps of downstream metabolites inside an intact subject by rationalizing the labeling of diet components with MRI-detectable isotopes (2H and 19F). We envision a new era of mapping and understanding the gut microbiome-cancer axis, which could be established upon successfully developing this tool. Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce a durable
Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce a durable

 Liquid biopsy is a promising non-invasive approach for early detection of cancer and monitoring disease progression and therapeutic response. We have recently developed a powerful and reliable method to isolate circulating small extracellular vesicles (sEVs) from plasma of breast cancer patients and showed that semi-quantitative proteomic profiling of sEVs can be used for early detection of breast cancer. Our preliminary analysis also suggests that the sEVs proteome could be used for prediction of recurrent disease. In this proof-of-concept project, we proposed not only to improve our isolation and profiling methods of circulating sEVs, but also to identify possible connections between the transcriptome of the primary breast tumors and the proteome of the circulating sEVs. To this end, we plan to profile matching samples of sEVs and primary tumors from breast cancer patients and to establish prediction models by machine learning as well as connectivity map between the tumor transcriptome and the sEVs proteome using computational methods. Deconvolution models will be applied to stratify sEVs based on their cells of origin. This combined analysis may predict sEVs biomarkers for distinct properties of the primary tumors, including tumor stage, tumor composition, heterogeneity, and response to treatment.
Liquid biopsy is a promising non-invasive approach for early detection of cancer and monitoring disease progression and therapeutic response. We have recently developed a powerful and reliable method to isolate circulating small extracellular vesicles (sEVs) from plasma of breast cancer patients and showed that semi-quantitative proteomic profiling of sEVs can be used for early detection of breast cancer. Our preliminary analysis also suggests that the sEVs proteome could be used for prediction of recurrent disease. In this proof-of-concept project, we proposed not only to improve our isolation and profiling methods of circulating sEVs, but also to identify possible connections between the transcriptome of the primary breast tumors and the proteome of the circulating sEVs. To this end, we plan to profile matching samples of sEVs and primary tumors from breast cancer patients and to establish prediction models by machine learning as well as connectivity map between the tumor transcriptome and the sEVs proteome using computational methods. Deconvolution models will be applied to stratify sEVs based on their cells of origin. This combined analysis may predict sEVs biomarkers for distinct properties of the primary tumors, including tumor stage, tumor composition, heterogeneity, and response to treatment. In cancer care, patients undergoing chemotherapy are required to manage their illness independently. Such management involves following the doctor's instructions and making critical decisions that affect their survival, such as reporting side effects or going to the emergency room. This research aimed to identify the types of knowledge patients need to manage their symptoms and side effects effectively. We focused on two types of knowledge: a) procedural knowledge, which involves understanding the steps to follow in various scenarios, and b) mechanistic knowledge, which pertains to understanding the underlying processes behind these procedures.
In cancer care, patients undergoing chemotherapy are required to manage their illness independently. Such management involves following the doctor's instructions and making critical decisions that affect their survival, such as reporting side effects or going to the emergency room. This research aimed to identify the types of knowledge patients need to manage their symptoms and side effects effectively. We focused on two types of knowledge: a) procedural knowledge, which involves understanding the steps to follow in various scenarios, and b) mechanistic knowledge, which pertains to understanding the underlying processes behind these procedures. Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce durable clinical
Immunotherapy has sparked new hope for oncology due to its remarkable ability to induce durable clinical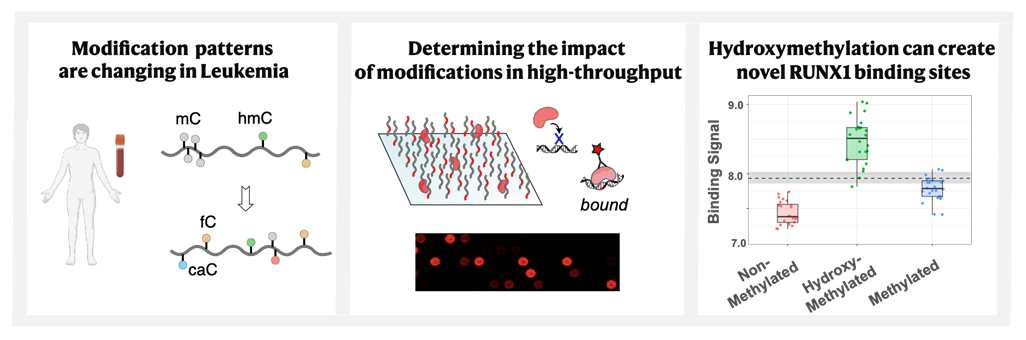 Epigenetic DNA modifications are continuously formed and removed in human cells, and disruptions in the activity of DNA-binding proteins that regulate these processes can lead to cancer development. We employed a cross-disciplinary approach, combining cancer genetics, high-throughput biophysics, and nucleic acid chemistry, to conduct a comprehensive analysis of cytosine modifications in healthy individuals and AML patients across different age groups. Through novel in vitro assays, we assessed the impact of these modifications on key transcription factors involved in disease progression. Our study presents the largest repository to date of quantitative data on how DNA modifications can profoundly alter the protein-DNA binding landscape, often redirecting oncogenic transcription factors such as RUNX1 and FOXO3A to non-target sequences. We anticipate that these findings will enhance our understanding of the principles governing modification formation and pave the way for a deeper molecular insight into AML development.
Epigenetic DNA modifications are continuously formed and removed in human cells, and disruptions in the activity of DNA-binding proteins that regulate these processes can lead to cancer development. We employed a cross-disciplinary approach, combining cancer genetics, high-throughput biophysics, and nucleic acid chemistry, to conduct a comprehensive analysis of cytosine modifications in healthy individuals and AML patients across different age groups. Through novel in vitro assays, we assessed the impact of these modifications on key transcription factors involved in disease progression. Our study presents the largest repository to date of quantitative data on how DNA modifications can profoundly alter the protein-DNA binding landscape, often redirecting oncogenic transcription factors such as RUNX1 and FOXO3A to non-target sequences. We anticipate that these findings will enhance our understanding of the principles governing modification formation and pave the way for a deeper molecular insight into AML development.