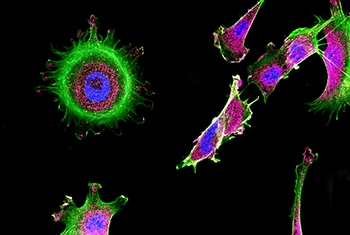
The Weizmann-Brazil cancer models unit support Weizmann scientists in selecting and developing the most suitable in vivo cancer models for their projects. We specialize in the establishment and maintenance of Patient-Derived Xenograft (PDX) and orthotopic tumor models that preserve key characteristics of human malignancies, including tumor heterogeneity and microenvironmental interactions, as well as common models.
PDX models generated by implanting freshly obtained samples from patients in the operating room after biopsy or tumor resection to immunodeficient animals resemble the original tumor growing within the human patient to the greatest possible extent. They can then be transferred from the first animal model to additional specimens, eventually propagating the tumors into a large cohort - all bearing the same human tumor. This cohort provides the researchers with an opportunity to perform a large variety of experimental manipulations, in parallel on identical replicas of the original tumor. These manipulations enable the identification of critical genes whose targeting can stop tumor growth or even kill the tumor.
Using PDX models in the Weizmann Cancer Models Unit yielded multiple publications. In a recent project in collaboration with the lab of Prof. Yossi Yarden lung PDX model were used to study the novel combination of bispecific antibodies to overcome drug resistance in EGFR mutation bearing tumors.
In a collaboration with Prof Rivka Dikstein a small-molecule inhibitors (1Ais) that disrupt eIF1A–RPS10 interactions was identified, revealing new regulatory roles in translation and demonstrating selective effects on cancer-related pathways. In this project, we used orthotopic ovarian cancer mouse models to demonstrate that 1Ais effectively reduces tumor growth without toxicity, highlighting their therapeutic potential.
Another project recently published by the group of Prof. Samuels generated SpotNeoMet, a pipeline that identifies clinically relevant neoantigens in therapy-resistant cancers, leading to the discovery of highly immunogenic TCRs targeting the AR H875Y mutation. These TCRs effectively eliminated prostate cancer cells in vitro and in vivo using mouse xenograft models, highlighting a promising therapeutic approach for metastatic castration-resistant prostate cancer.
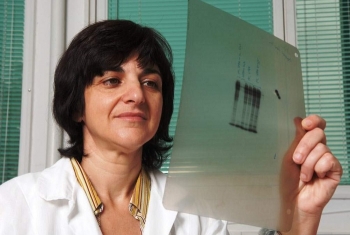
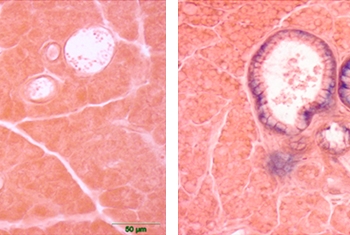
Picture (left): Preparation of a tumor sample extracted from a PDX mouse, prior to implantation into additional animal models.